Difference between revisions of "MITK-nnInteractive"
| Line 31: | Line 31: | ||
== Basic Usage == | == Basic Usage == | ||
| + | |||
| + | [[File:NnInteractive button in Segmentation plugin.png|thumb]] | ||
# Load an Image | # Load an Image | ||
Revision as of 21:06, 11 March 2025
Overview and Download
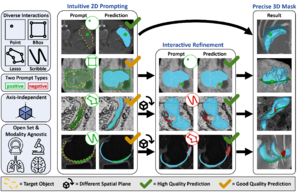
In this preview, nnInteractive, the universal 3-d promptable segmentation model, is integrated into MITK. It enables interactive, open-set segmentation using various prompts such as points, scribbles, boxes, and lasso. The model translates intuitive 2-d interactions into full 3-d segmentations, facilitating efficient and accurate biomedical image analysis.
Download: MITK nnInteractive Preview 1 for Windows
Note that this is an experimental preview version of MITK that is potentially unstable. nnInteractive will be fully integrated for Windows and Linux in the next regular MITK release (ETA: June 2025). We are in active exchange with the creators of nnInteractive to evaluate options for macOS support, which is currently natively unsupported by nnInteractive.
If you already want to check out nnInteractive on Linux, we recommend the napari integration.
Features
- Designed for medical imaging: Tailored for modalities like MRI, CT, and microscopy, ensuring accurate tissue and structure segmentation
- Multi-modal prompt support: Points, scribbles, bounding boxes, and lasso
- Full 3-d segmentation from simple 2-d interactions
- Multi-plane interaction: Supports user interaction across axial, sagittal, and coronal planes for precise anatomical structure delineation.
- 3-d visualization: Provides 3-d rendering of segmentations for enhanced spatial understanding and validation.
Demo videos
Check out the nnInteractive napari integration for demo videos showcasing nnInteractive in action. This integration serves as an alternative to the MITK nnInteractive Preview, especially for Linux users, as our MITK integration does not yet support Linux. Full Linux support is expected in the next MITK release (ETA: June 2025).
Installation
- Download the latest MITK nnInteractive Preview ZIP archive from above.
- No installation needed, just unzip and run MitkWorkbench.bat.
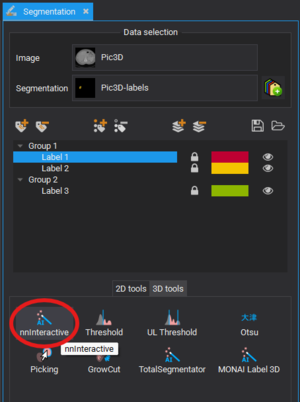
Basic Usage
- Load an Image
- Open the MITK Workbench and load your 3-d medical image (e.g. MRI, CT, microscopy images).
- Select the nnInteractive tool
- Open the Segmentation plugin.
- Create a segmentation.
- Click on nnInteractive under 3D tools.
Workflow
- Click on the "Initialize" button to get the nnInteractive backend ready for a segmentation session.
- The following interactions/prompts are available after a successful initialization:
- Point: Click on the image to place points.
- Box: Click and drag on the image to place a box resp. rectangle.
- Scribble: Click and move the mouse cursor over the image to scribble.
- Lasso: Click and move the mouse to outline a region of interest.
- Initialize with Mask: Use an existing segmentation label as initial mask for further refinement.
You can combine any interaction type and individually decide if an interaction should be positive or negative. The initialization with a mask will reset all previous uncommitted interactions.
Don't forget to click on "Confirm Segmentation" to finally commit the shown preview into a segmentation label.
Save Results
Once your segmentation is complete you can save your work by right-clicking on your segmentation in the Data Manager (typically on the left) and selecting "Save...". This will open a file dialog where you can export segmentations in supported formats like NRRD for further analysis or sharing. Note that saving segmentation as DICOM SEGs is only supported if the reference image was also loaded from DICOM files.
Performance Considerations
nnInteractive is a deep learning-based model that requires significant computational resources. To ensure optimal performance we highly recommend a CUDA-enabled GPU with tensor cores and at least 6 GBs of VRAM like an NVIDIA GeForce RTX 2060 or better. Performance on older GPUs without tensor cores like the NVIDIA GeForce GTX 1080 will already take twice as long during interaction. Make sure to use an up-to-date graphics driver.
As a last resort, nnInteractive can also run on CPUs but with significant (!) performance drops of at least a magnitude in comparision to a supported GPU.
Contributions and Feedback
Contributions and feedback are welcome! Feel free to report issues or suggest improvements via GitHub.
License
- MITK Workbench: BSD-3-Clause license
- nnInteractive: Apache 2.0 license
- nnInteractive model weights: CC-BY-NC-SA 4.0